
Acronym ML stands for maximum likelihood.
ML estimators are calculated in such a way that they maximize the likelihood of n independent observations. That is, if f(xi;θ1,…,θp) is the value of the density function for the i-th observation, which depends on parameters θ1,…,θp, the function to be maximized is L(θ1,…,θp;x1,…,xn)=n∏i=1f(xi;θ1,…,θp). The set of numbers ˆθ1,…,ˆθp, such that they maximize L, are the so called maximum likelihood estimators.
First, we load package distrib,
load("distrib")$
Given a normal, or gaussian, population with unknown parameters μ and σ, once we have an independent sample of size n, (x1,x2,…,xn), our likelihood function takes the form
L: product(pdf_normal(x[i],mu,sigma), i, 1, n);
n∏i=1e−(xi−μ)22σ2√2√πσ
Taking the logarithm of the above expression, we get the loglikelihood function,
logL: ev(log(L), logexpand=all);
n∑i=1(−logσ−(xi−μ)22σ2−logπ2−log22)
As we want to maximize the likelihood, or equivalently the loglikelihood, we need to equal to zero the partial derivatives of the loglikelihood with respect to μ and σ. The derivative with respect to μ is
logLmu: ev(diff(logL, mu), simpsum=true);
∑ni=1xi−μnσ2
Equating to zero and isolating μ yields
hatmu: solve(logLmu, mu);
[μ=∑ni=1xin]
Which we recognize as the sample mean, so that our result can be summarized as ˆμ=ˉx.
We repeat a similar procedure for σ2,
logLsi: ev(diff(logL, sigma), simpsum=true) $ hatsi: solve(logLsi, sigma^2);
[σ2=∑ni=1(xi−μ)2n]
Which is the sample variance, so that ˆσ2=s2.
The expression for the variance can be written only in terms of the sample values,
subst(hatmu, hatsi);
[σ2=∑ni=1(xi−∑ni=1xin)2n]
Let us now proceed with the numerical part of this session. Instead of taking our sample from a real world population, we can make a simulation calling function random_normal from package distrib. Let n=50 be the sample size, μ=50, and σ=2.5,
(n: 50, mu: 50, sigma: 2.5) $ fpprintprec: 4 $ x: random_normal(mu, sigma, n) ;
[53.21,50.45,52.16,51.37,44.37,52.65,50.92,51.25,52.18,48.49,52.49,46.19,47.32,47.73,50.58,48.6,49.21,52.32,49.95,48.7,55.38,51.36,48.3,49.34,52.6,48.8,47.52,44.53,49.25,44.47,48.58,48.33,48.39,49.36,43.98,53.73,51.93,51.76,46.22,53.61,51.2,50.16,54.3,46.86,56.86,49.21,50.75,45.22,53.87,48.85]
In order to calculate the sample mean and variance, we need to load the descriptive package. We also include the sample standard deviation,
load("descriptive") $
ml: [mean(x), var(x), std(x)];
[49.9,8.529,2.92]
As a result, ˆμ=49.9, and ˆσ=2.92. Remember that our sample was artificially drawn from a normal population with parametrrs μ=50, and σ=2.5. Also remember that the ML estimator of the variance is a biased estimator.
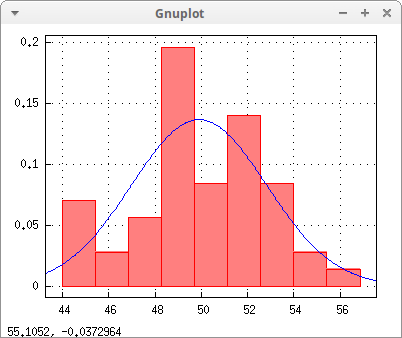
With help of function histogram, also implemented in package descriptive, we can show a graphical representation of the sample together with the estimated gaussian probability density,
[m, s]: [ml[1], ml[3]] $
draw2d(
grid = true,
dimensions = [400,300],
histogram_description(
x,
nclasses = 9,
frequency = density,
fill_density = 0.5),
explicit(pdf_normal(z,m,s), z, m - 3*s, m + 3* s) ) $
© 2017, TecnoStats.