
The Weibull distribution is extensively used in survival and reliability analysis to model the waiting time for an event to occur. It depends on two positive parameters a and b,
load("distrib")$
pdf_weibull(t, a, b);
a(tb)a−1e−(tb)ab
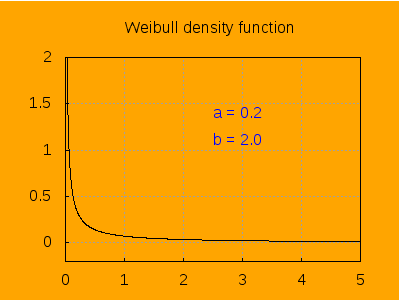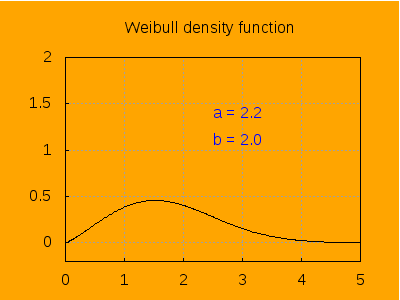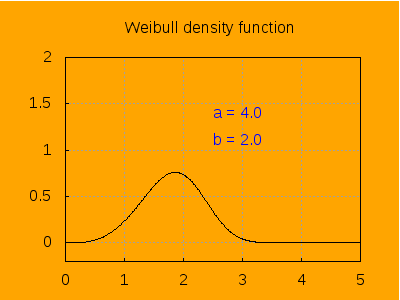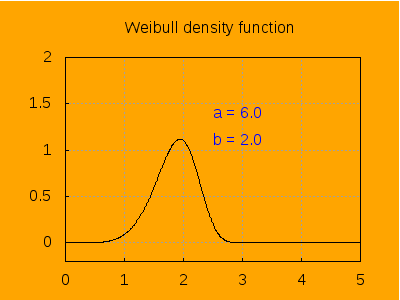
This is the aspect of the density function for different values of a, keeping b=2.
load("draw")$
b: 2 $
fpprintprec: 4 $
draw(
terminal = animated_gif,
delay = 20,
dimensions = [400, 300],
makelist(
gr2d(
title = "Weibull density function",
background_color = orange,
grid = true,
yrange = [-0.2, 2],
label_alignment = left,
label([string('a = a), 2.5, 1.4]),
label([string('b = 2.0), 2.5, 1.1]),
color = black,
explicit(pdf_weibull(t, a, b), t, 0, 5)),
a, 0.2 , 6, 0.2) ) $
In reliability and survival analysis, the survival function, also known as reliability function in the industrial context, is defined as S(t)=Pr(T>t)=1−F(t),
S(t,a,b) := ''(1 - cdf_weibull(t, a, b));
S(t,a,b):=e−(tb)a
Given a sample (t1,t2,…,tn) with uncensored and right-censored data, the Kaplan-Meier estimator for the survival function is ˆS(t)=∏tj≤t(1−djnj), where dj is the number of dying subjects at time tj, and nj is the number of subjects still alive just before time tj. An implementation of this algorithm follows,
/* Kaplan-Meier estimator */
load("descriptive")$
charfun2(z,l1,l2):= charfun(l1 <= z and z < l2)$
km(x) := block([s,ss,t,st,n,d,c,i:2,idx:1,hatS],
s: sort(x),
ss: length(x),
t: append([0],
listify(
setify(
first(
transpose(
subsample(s,lambda([z], second(z)=1),1)))))),
st: length(t),
n: makelist(0,k,st),
d: makelist(0,k,st),
c: makelist(0,k,st),
n[1]: ss,
while i <= st do
( n[i]: n[i-1] - d[i-1] - c[i-1],
while idx <= ss and s[idx][1] <= t[i] do
( if s[idx][2] = 1
then d[i]: d[i] + 1
elseif s[idx][1] < t[i]
then n[i]: n[i] - 1
else c[i]: c[i] + 1,
idx: idx + 1),
i: i + 1 ),
hatS: 1- d / n,
for k:2 thru st do
hatS[k]: hatS[k-1] * hatS[k],
sum(charfun2(z,t[i-1],t[i]) * hatS[i-1], i, 2, st) +
charfun2(z,t[st],inf) * hatS[st] ) $
In order to see the Kaplan-Meier estimator in action, let's simulate a random sample of size 15 drawn from a Weibull distribution with parameters a=10 and b=7. Together with the time values we include a number to indicate whether the time is censored (0) or not (1). We consider observations longer than 8 to be right censored,
fpprintprec:4 $ /* for pretty printing */ (a: 10, b: 7, c: 8) $ x: random_weibull(a, b, 15) $ x: map(lambda([z], if z < c then [z,1] else [8,0]), x);
[[7.578,1],[4.671,1],[7.163,1],[6.658,1],[8,0],[6.905,1],[6.248,1],[5.441,1],[6.05,1],[6.045,1],[7.158,1],[7.158,1],[7.566,1],[6.001,1],[5.537,1]]
And here is the estimated survival step function,
hatS(z) := ''(km(x)) ;
2charfun(7.566≤z∧z<7.578)15+charfun(7.163≤z∧z<7.566)5+4charfun(7.158≤z∧z<7.163)15+charfun(7.158≤z∧z<7.158)3+2charfun(6.905≤z∧z<7.158)5+7charfun(6.658≤z∧z<6.905)15+8charfun(6.248≤z∧z<6.658)15+3charfun(6.05≤z∧z<6.248)5+2charfun(6.045≤z∧z<6.05)3+11charfun(6.001≤z∧z<6.045)15+4charfun(5.537≤z∧z<6.001)5+13charfun(5.441≤z∧z<5.537)15+14charfun(4.671≤z∧z<5.441)15+charfun(0≤z∧z<4.671)+charfun(7.578≤z)15
As an example, let's calculate the probability of a subject to be still alive at time t=7,
hatS(7);
25
And a plot of it,
draw2d( terminal = png, grid = true, background_color = orange, dimensions = [400, 300], yrange = [0, 1], title = "Kaplan-Meier estimator", line_width = 3, explicit(hatS(z), z, 0, 8), color = white, line_width = 1, explicit(S(z, a, b), z, 0, 8)) $
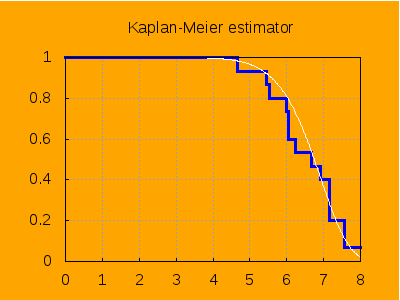
We have plotted in white color the true survival function, which we know, since our sample have been generated by simulation.
Let's now try to estimate parameters a and b from the sample. One of the standard methods is via the maximum likelihood. But in this case we'll use a method based on linear regression. See the following result,
kill(a,b)$ 'S(z,a,b) = S(z,a,b);
S(z,a,b)=e−(zb)a
log(lhs(%)) = log(rhs(%));
logS(z,a,b)=−(zb)a
-lhs(%) = -rhs(%);
−logS(z,a,b)=(zb)a
log(lhs(%)) = ev(log(rhs(%)), logexpand = all);
log(−logS(z,a,b))=a(logz−logb)
lhs(%) = expand(rhs(%));
log(−logS(z,a,b))=alogz−alogb
According to this last result, there is a linear relationship between logz and log(−logS(z,a,b)). We can estimate parameters m=a and n=−alogb by simple linear regression and then obtain estimators for a and b.
We extract from the sample true life times,
x1: subsample(x, lambda([v], second(v)=1),1);
(7.5784.6717.1636.6586.9056.2485.4416.056.0457.1587.1587.5666.0015.537)
Estimate the survival time for each observation,
y1: float(matrixmap(hatS,x1));
(0.066670.93330.20.46670.40.53330.86670.60.66670.26670.33330.13330.73330.8)
Build a table with the two lists, removing those pairs with zero survival probability to avoid problems with logaritms. This task is performed with function subsample defined in package descriptive,
w: subsample(addcol(x1,y1), lambda([v], second(v)#0.0),1,2);
(7.5780.066674.6710.93337.1630.26.6580.46676.9050.46.2480.53335.4410.86676.050.66.0450.66677.1580.26677.1580.33337.5660.13336.0010.73335.5370.8)
Now we transformate this matrix to obtain pairs of the form (logz,log(−logˆS(z))). Function transform_sample, also from package descriptive, is our friend here,
pts: transform_sample(w, [a,b], [log(a),log(-log(b))]) $
(2.0250.99621.541−2.6741.9690.47591.896−0.27161.932−0.087421.832−0.46421.694−1.9441.8−0.67171.799−0.90271.9680.2791.9680.094052.0240.70061.792−1.1711.711−1.5)
We are now ready for a least square adjustment,
load (lsquares)$
le: float(lsquares_estimates(pts,
[u,v],
v=m*u+n,
[m,n],
initial=[3,3],
iprint=[-1,0]));
[[m=7.315,n=−14.07]]
Since m=a and n=−alogb, the estimator for a is
aa: rhs(le[1][1]) ;
7.315
And for b,
float(solve(−aa*log(b)= rhs(le[1][2])));
[b=6.845]
Remember that our sample was obtained by simulation with a=10 and b=7.
© 2017, TecnoStats.